A Gold-Containing Drug Against Parasitic Polyamine Metabolism: The X-Ray Structure of Trypanothione Reductase from Leishmania Infantum in Complex with Auranofin Reveals a Dual Mechanism of Enzyme Inhibition.
Ilari, A., Baiocco, P., Messori, L., Fiorillo, A., Boffi, A., Gramiccia, M., Di Muccio, T., Colotti, G.(2012) Amino Acids 42: 803
- PubMed: 21833767
- DOI: https://doi.org/10.1007/s00726-011-0997-9
- Primary Citation of Related Structures:
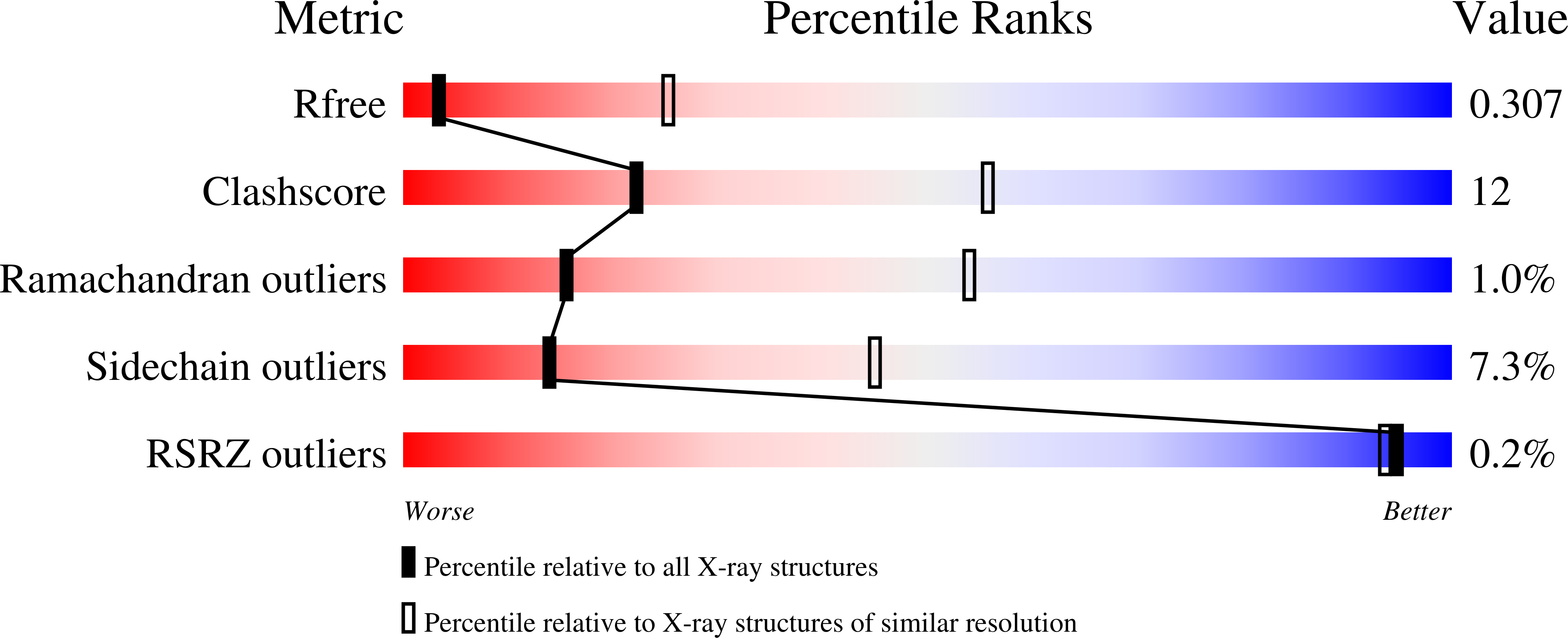
2YAU - PubMed Abstract:
Auranofin is a gold(I)-containing drug in clinical use as an antiarthritic agent. Recent studies showed that auranofin manifests interesting antiparasitic actions very likely arising from inhibition of parasitic enzymes involved in the control of the redox metabolism. Trypanothione reductase is a key enzyme of Leishmania infantum polyamine-dependent redox metabolism, and a validated target for antileishmanial drugs. As trypanothione reductase contains a dithiol motif at its active site and gold(I) compounds are known to be highly thiophilic, we explored whether auranofin might behave as an effective enzyme inhibitor and as a potential antileishmanial agent. Notably, enzymatic assays revealed that auranofin causes indeed a pronounced enzyme inhibition. To gain a deeper insight into the molecular basis of enzyme inhibition, crystals of the auranofin-bound enzyme, in the presence of NADPH, were prepared, and the X-ray crystal structure of the auranofin-trypanothione reductase-NADPH complex was solved at 3.5 Å resolution. In spite of the rather low resolution, these data were of sufficient quality as to identify the presence of the gold center and of the thiosugar of auranofin, and to locate them within the overall protein structure. Gold binds to the two active site cysteine residues of TR, i.e. Cys52 and Cys57, while the thiosugar moiety of auranofin binds to the trypanothione binding site; thus auranofin appears to inhibit TR through a dual mechanism. Auranofin kills the promastigote stage of L. infantum at micromolar concentration; these findings will contribute to the design of new drugs against leishmaniasis.
Organizational Affiliation:
Department of Biochemical Sciences, Institute of Molecular Biology and Pathology CNR, Sapienza University of Rome, P.le A. Moro 5, 00185, Rome, Italy. [email protected]