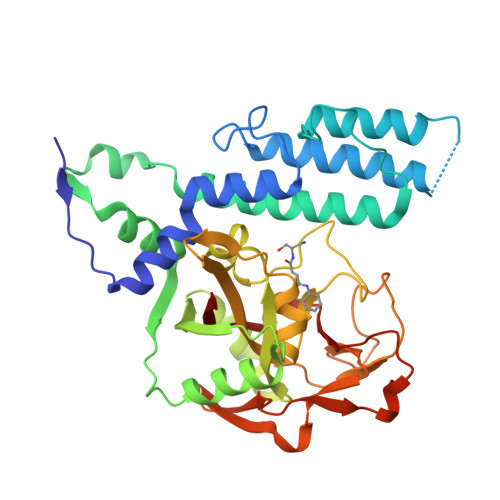
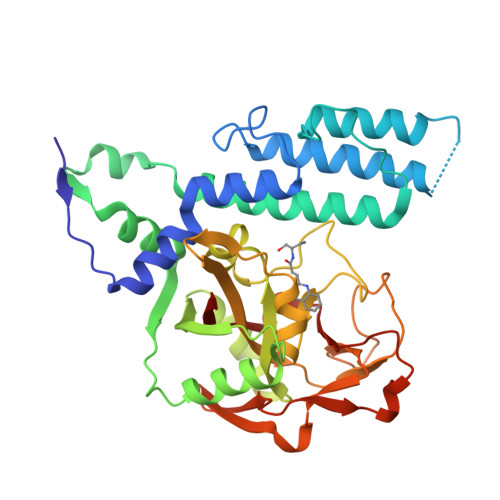
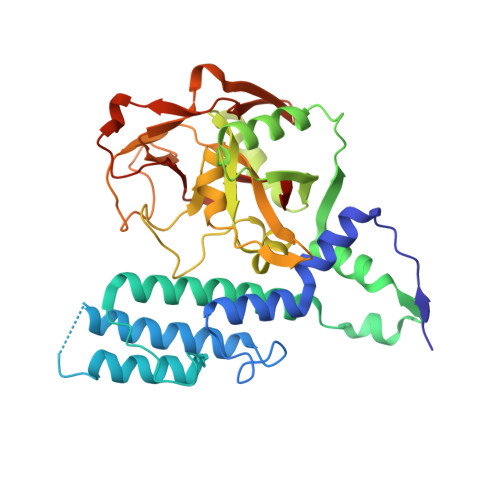
Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
Lindgren, A.E., Karlberg, T., Ekblad, T., Spjut, S., Thorsell, A.G., Andersson, C.D., Nhan, T.T., Hellsten, V., Weigelt, J., Linusson, A., Schuler, H., Elofsson, M.(2013) J Med Chem 56: 9556-9568
- PubMed: 24188023
- DOI: https://doi.org/10.1021/jm401394u
- Primary Citation of Related Structures:
4L6Z, 4L70, 4L7L, 4L7N, 4L7O, 4L7P, 4L7R, 4L7U - PubMed Abstract:
The racemic 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[1-(pyridin-2-yl)ethyl]propanamide, 1, has previously been identified as a potent but unselective inhibitor of diphtheria toxin-like ADP-ribosyltransferase 3 (ARTD3). Herein we describe synthesis and evaluation of 55 compounds in this class. It was found that the stereochemistry is of great importance for both selectivity and potency and that substituents on the phenyl ring resulted in poor solubility. Certain variations at the meso position were tolerated and caused a large shift in the binding pose. Changes to the ethylene linker that connects the quinazolinone to the amide were also investigated but proved detrimental to binding. By combination of synthetic organic chemistry and structure-based design, two selective inhibitors of ARTD3 were discovered.
Organizational Affiliation:
Department of Chemistry, Umeå University , SE-90187 Umeå, Sweden.