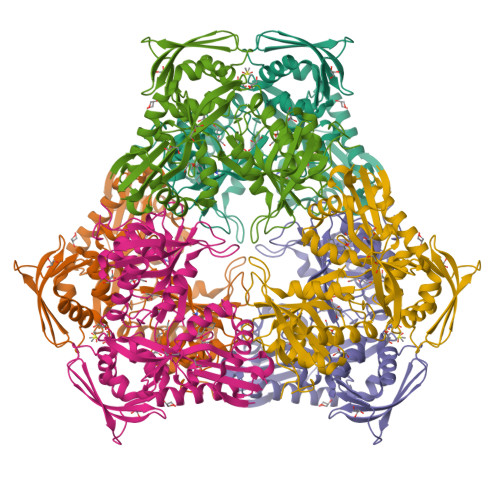
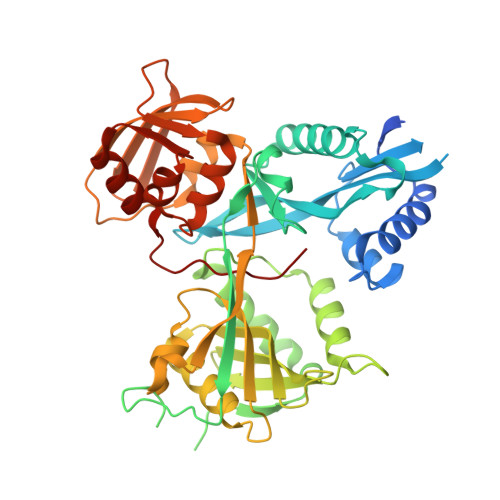
Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Pang, A.H., Green, K.D., Punetha, A., Thamban Chandrika, N., Howard, K.C., Garneau-Tsodikova, S., Tsodikov, O.V.(2023) Biochemistry 62: 710-721
- PubMed: 36657084
- DOI: https://doi.org/10.1021/acs.biochem.2c00658
- Primary Citation of Related Structures:
8F4A, 8F4T, 8F4U, 8F4W, 8F4Z, 8F51, 8F55, 8F57, 8F58 - PubMed Abstract:
Over one and a half million people die of tuberculosis (TB) each year. Multidrug-resistant TB infections are especially dangerous, and new drugs are needed to combat them. The high cost and complexity of drug development make repositioning of drugs that are already in clinical use for other indications a potentially time- and money-saving avenue. In this study, we identified among existing drugs five compounds: azelastine, venlafaxine, chloroquine, mefloquine, and proguanil as inhibitors of acetyltransferase Eis from Mycobacterium tuberculosis , a causative agent of TB. Eis upregulation is a cause of clinically relevant resistance of TB to kanamycin, which is inactivated by Eis-catalyzed acetylation. Crystal structures of these drugs as well as chlorhexidine in complexes with Eis showed that these inhibitors were bound in the aminoglycoside binding cavity, consistent with their established modes of inhibition with respect to kanamycin. Among three additionally synthesized compounds, a proguanil analogue, designed based on the crystal structure of the Eis-proguanil complex, was 3-fold more potent than proguanil. The crystal structures of these compounds in complexes with Eis explained their inhibitory potencies. These initial efforts in rational drug repositioning can serve as a starting point in further development of Eis inhibitors.
Organizational Affiliation:
Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, Kentucky40536-0596, United States.