Bornyl Diphosphate Synthase: Structure and Strategy for Carbocation Manipulation by a Terpenoid Cyclase
Whittington, D.A., Wise, M.L., Urbansky, M., Coates, R.M., Croteau, R.B., Christianson, D.W.(2002) Proc Natl Acad Sci U S A 99: 15375-15380
- PubMed: 12432096
- DOI: https://doi.org/10.1073/pnas.232591099
- Primary Citation of Related Structures:
1N1B, 1N1Z, 1N20, 1N21, 1N22, 1N23, 1N24 - PubMed Abstract:
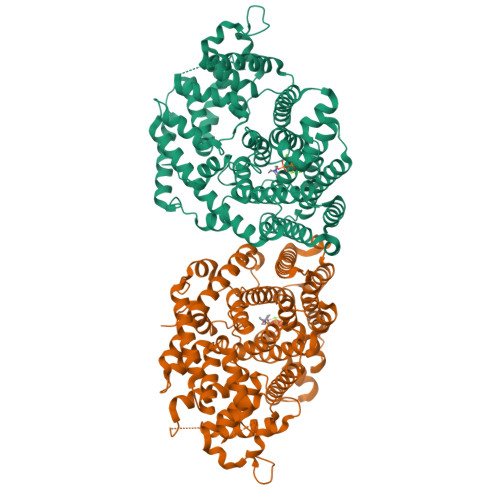
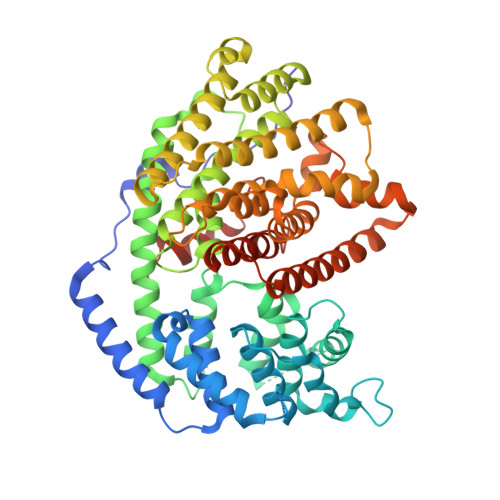
The x-ray crystal structure of dimeric (+)-bornyl diphosphate synthase, a metal-requiring monoterpene cyclase from Salvia officinalis, is reported at 2.0-A resolution. Each monomer contains two alpha-helical domains: the C-terminal domain catalyzes the cyclization of geranyl diphosphate, orienting and stabilizing multiple reactive carbocation intermediates; the N-terminal domain has no clearly defined function, although its N terminus caps the active site in the C-terminal domain during catalysis. Structures of complexes with aza analogues of substrate and carbocation intermediates, as well as complexes with pyrophosphate and bornyl diphosphate, provide "snapshots" of the terpene cyclization cascade.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia 19104-6323, USA.