Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Thiel, P., Roglin, L., Meissner, N., Hennig, S., Kohlbacher, O., Ottmann, C.(2013) Chem Commun (Camb) 49: 8468-8470
- PubMed: 23939230
- DOI: https://doi.org/10.1039/c3cc44612c
- Primary Citation of Related Structures:
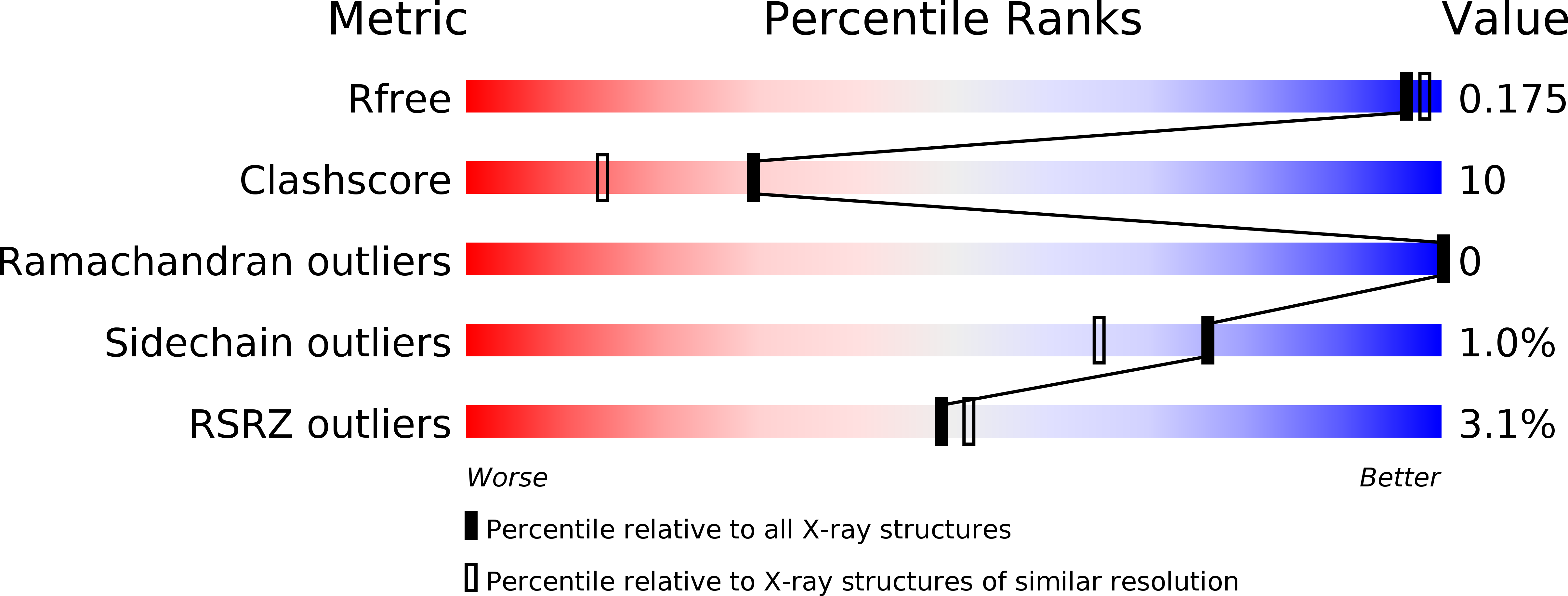
3T0L, 3T0M, 4DHM, 4DHN, 4DHO, 4DHP, 4DHQ, 4DHR, 4DHS, 4DHT, 4DHU - PubMed Abstract:
We report first non-covalent and exclusively extracellular inhibitors of 14-3-3 protein-protein interactions identified by virtual screening. Optimization by crystal structure analysis and in vitro binding assays yielded compounds capable of disrupting the interaction of 14-3-3σ with aminopeptidase N in a cellular assay.
Organizational Affiliation:
Chemical Genomics Centre of the Max Planck Society, Otto-Hahn-Straße 15, D-44227 Dortmund, Germany.