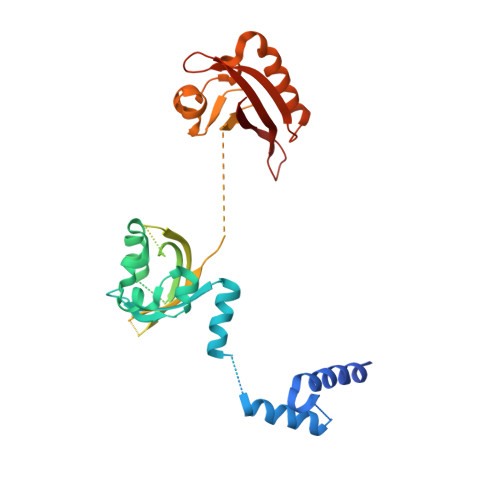
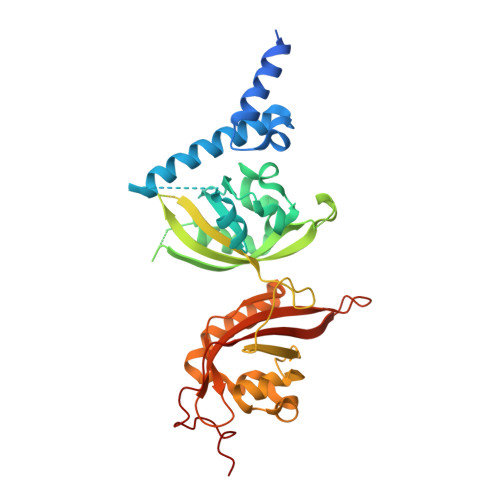
Structural integration in hypoxia-inducible factors.
Wu, D., Potluri, N., Lu, J., Kim, Y., Rastinejad, F.(2015) Nature 524: 303-308
- PubMed: 26245371
- DOI: https://doi.org/10.1038/nature14883
- Primary Citation of Related Structures:
4ZP4, 4ZPH, 4ZPK, 4ZPR, 4ZQD - PubMed Abstract:
The hypoxia-inducible factors (HIFs) coordinate cellular adaptations to low oxygen stress by regulating transcriptional programs in erythropoiesis, angiogenesis and metabolism. These programs promote the growth and progression of many tumours, making HIFs attractive anticancer targets. Transcriptionally active HIFs consist of HIF-α and ARNT (also called HIF-1β) subunits. Here we describe crystal structures for each of mouse HIF-2α-ARNT and HIF-1α-ARNT heterodimers in states that include bound small molecules and their hypoxia response element. A highly integrated quaternary architecture is shared by HIF-2α-ARNT and HIF-1α-ARNT, wherein ARNT spirals around the outside of each HIF-α subunit. Five distinct pockets are observed that permit small-molecule binding, including PAS domain encapsulated sites and an interfacial cavity formed through subunit heterodimerization. The DNA-reading head rotates, extends and cooperates with a distal PAS domain to bind hypoxia response elements. HIF-α mutations linked to human cancers map to sensitive sites that establish DNA binding and the stability of PAS domains and pockets.
Organizational Affiliation:
Metabolic Disease Program, Sanford Burnham Prebys Medical Discovery Institute, Orlando, Florida 32827, USA.