Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Engilberge, S., Riobe, F., Wagner, T., Di Pietro, S., Breyton, C., Franzetti, B., Shima, S., Girard, E., Dumont, E., Maury, O.(2018) Chemistry 24: 9739-9746
- PubMed: 29806881
- DOI: https://doi.org/10.1002/chem.201802172
- Primary Citation of Related Structures:
6F2F, 6F2H, 6F2I, 6F2J, 6F2K, 6F2M, 6FRM, 6FRN, 6FRO, 6FRQ - PubMed Abstract:
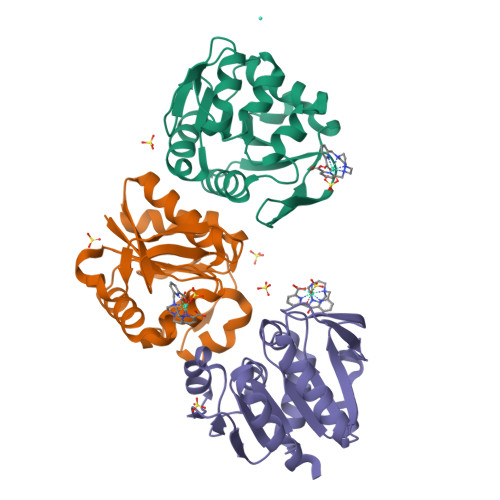
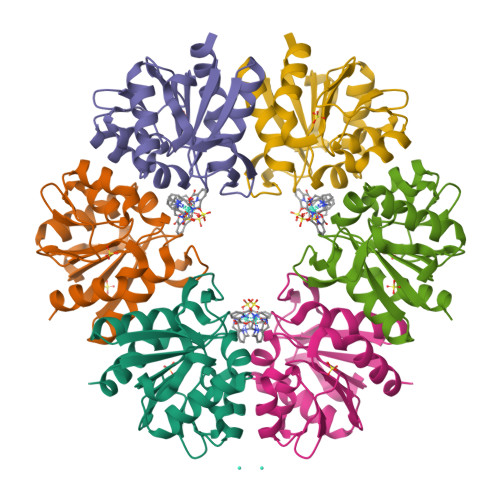
Crystallophores are lanthanide complexes that act as powerful auxiliary for protein crystallography due to their strong nucleating and phasing effects. To get first insights on the mechanisms behind nucleation induced by Crystallophore, we systematically identified various elaborated networks of supramolecular interactions between Tb-Xo4 and subset of 6 protein structures determined by X-ray diffraction in complex with terbium-Crystallophore (Tb-Xo4). Such interaction mapping analyses demonstrate the versatile binding behavior of the Crystallophore and pave the way to a better understanding of its unique properties.
Organizational Affiliation:
Univ Grenoble Alpes, CEA, CNRS, IBS, 38000, Grenoble, France.