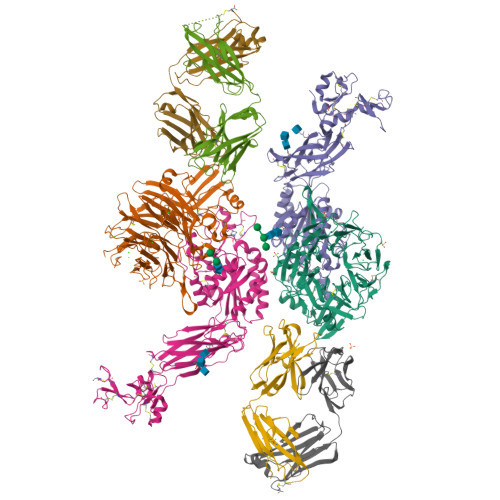
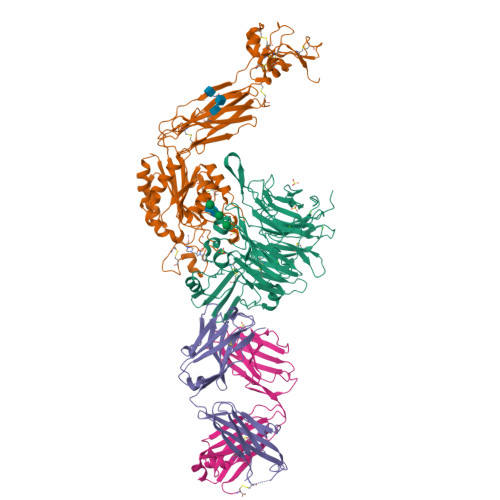
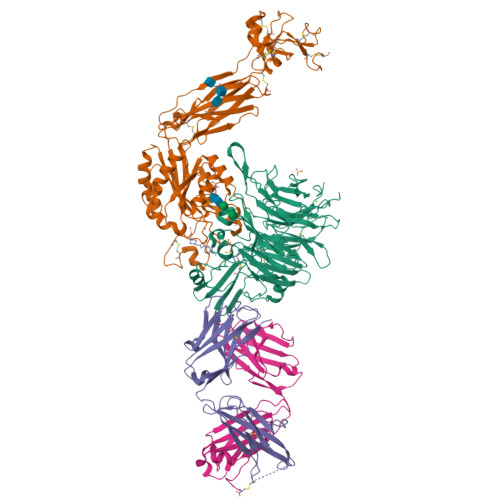
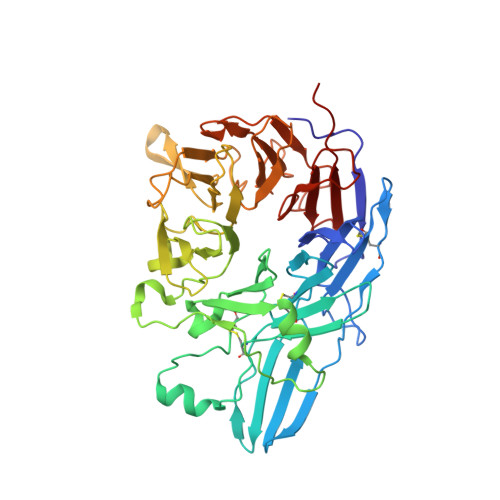
A general chemical principle for creating closure-stabilizing integrin inhibitors.
Lin, F.Y., Li, J., Xie, Y., Zhu, J., Huong Nguyen, T.T., Zhang, Y., Zhu, J., Springer, T.A.(2022) Cell 185: 3533-3550.e27
- PubMed: 36113427
- DOI: https://doi.org/10.1016/j.cell.2022.08.008
- Primary Citation of Related Structures:
7L8P, 7TCT, 7TD8, 7THO, 7TMZ, 7TPD, 7U60, 7U9F, 7U9V, 7UBR, 7UCY, 7UDG, 7UDH, 7UE0, 7UFH, 7UH8, 7UJE, 7UJK, 7UK9, 7UKO, 7UKP, 7UKT - PubMed Abstract:
Integrins are validated drug targets with six approved therapeutics. However, small-molecule inhibitors to three integrins failed in late-stage clinical trials for chronic indications. Such unfavorable outcomes may in part be caused by partial agonism, i.e., the stabilization of the high-affinity, extended-open integrin conformation. Here, we show that the failed, small-molecule inhibitors of integrins αIIbβ3 and α4β1 stabilize the high-affinity conformation. Furthermore, we discovered a simple chemical feature present in multiple αIIbβ3 antagonists that stabilizes integrins in their bent-closed conformation. Closing inhibitors contain a polar nitrogen atom that stabilizes, via hydrogen bonds, a water molecule that intervenes between a serine residue and the metal in the metal-ion-dependent adhesion site (MIDAS). Expulsion of this water is a requisite for transition to the open conformation. This change in metal coordination is general to integrins, suggesting broad applicability of the drug-design principle to the integrin family, as validated with a distantly related integrin, α4β1.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Program in Cellular and Molecular Medicine, Boston Children's Hospital, Harvard Medical School, Boston, MA 02115, USA.