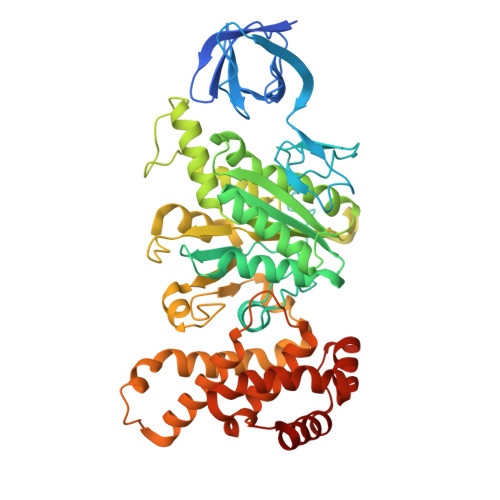
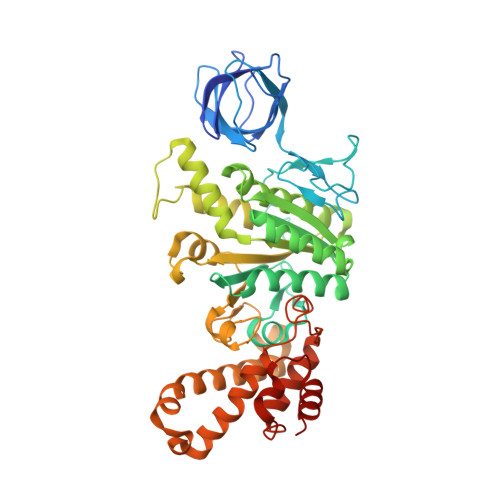
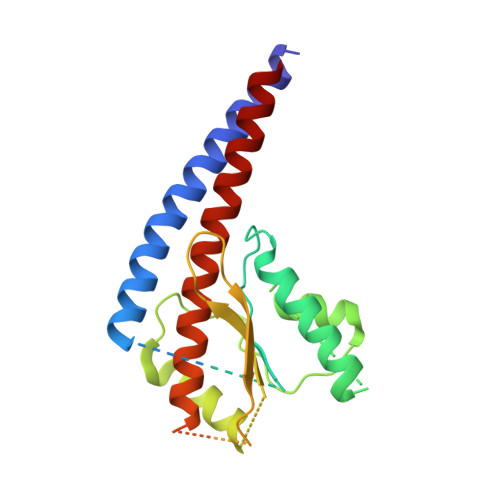
The molecular structure of an axle-less F 1 -ATPase.
Furlong, E.J., Reininger-Chatzigiannakis, I.P., Zeng, Y.C., Brown, S.H.J., Sobti, M., Stewart, A.G.(2024) Biochim Biophys Acta Bioenerg 1866: 149521-149521
- PubMed: 39428050
- DOI: https://doi.org/10.1016/j.bbabio.2024.149521
- Primary Citation of Related Structures:
8U1H, 9AVJ - PubMed Abstract:
F 1 F o ATP synthase is a molecular rotary motor that can generate ATP using a transmembrane proton motive force. Isolated F 1 -ATPase catalytic cores can hydrolyse ATP, passing through a series of conformational states involving rotation of the central γ rotor subunit and the opening and closing of the catalytic β subunits. Cooperativity in F 1 -ATPase has long thought to be conferred through the γ subunit, with three key interaction sites between the γ and β subunits being identified. Single molecule studies have demonstrated that the F 1 complexes lacking the γ axle still "rotate" and hydrolyse ATP, but with less efficiency. We solved the cryogenic electron microscopy structure of an axle-less Bacillus sp. PS3 F 1 -ATPase. The unexpected binding-dwell conformation of the structure in combination with the observed lack of interactions between the axle-less γ and the open β subunit suggests that the complete γ subunit is important for coordinating efficient ATP binding of F 1 -ATPase.
Organizational Affiliation:
Molecular, Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Darlinghurst, Australia; Division of Biomedical Science and Biochemistry, Research School of Biology, Australian National University, Acton, ACT, Australia.