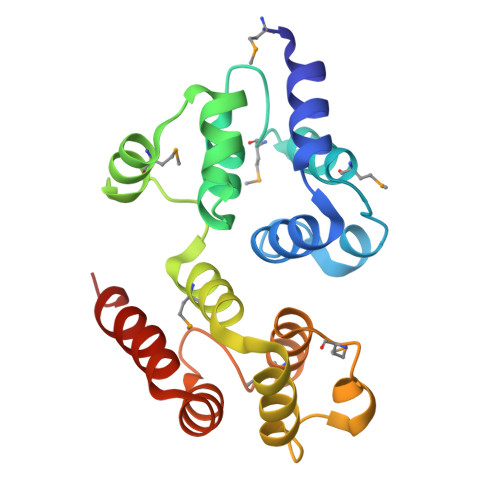
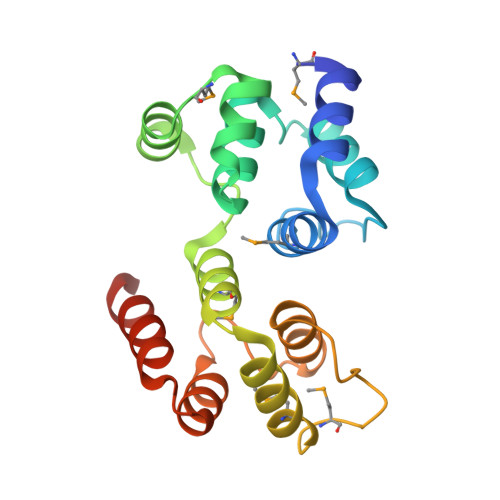
Reverse hierarchical DED assembly in the cFLIP-procaspase-8 and cFLIP-procaspase-8-FADD complexes.
Yang, C.Y., Tseng, Y.C., Tu, Y.F., Kuo, B.J., Hsu, L.C., Lien, C.I., Lin, Y.S., Wang, Y.T., Lu, Y.C., Su, T.W., Lo, Y.C., Lin, S.C.(2024) Nat Commun 15: 8974-8974
- PubMed: 39419969
- DOI: https://doi.org/10.1038/s41467-024-53306-1
- Primary Citation of Related Structures:
8YM4, 8YM5, 8YM6, 8YNI, 8YNK, 8YNL, 8YNM, 8YNN - PubMed Abstract:
cFLIP, a master anti-apoptotic regulator, targets the FADD-induced DED complexes of procaspase-8 in death receptor and ripoptosome signaling pathways. Several tumor cells maintain relatively high levels of cFLIP in achieving their immortality. However, understanding the three-dimensional regulatory mechanism initiated or mediated by elevated levels of cFLIP has been limited by the absence of the atomic coordinates for cFLIP-induced DED complexes. Here we report the crystal plus cryo-EM structures to uncover an unconventional mechanism where cFLIP and procaspase-8 autonomously form a binary tandem DED complex, independent of FADD. This complex gains the ability to recruit FADD, thereby allosterically modulating cFLIP assembly and partially activating caspase-8 for RIPK1 cleavage. Our structure-guided mutagenesis experiments provide critical insights into these regulatory mechanisms, elucidating the resistance to apoptosis and necroptosis in achieving immortality. Finally, this research offers a unified model for the intricate bidirectional hierarchy-based processes using multiprotein helical assembly to govern cell fate decisions.
Organizational Affiliation:
Genomics Research Center, Academia Sinica, Taipei, 11529, Taiwan.